The MIVIA LDGraphs (MIVIA Large Dense Graphs) dataset is a new dataset for benchmarking exact graph matching algorithms. It aims to extend the MIVIA graphs dataset, widely used in the last ten years, with bigger and more dense graphs, so as to face with the problems nowadays encountered in real applications devoted for instance to bioinformatics and social network analysis.
If you are using this library please cite:
The MIVIA graphs dataset
The MIVIA graphs dataset has been one of the mostly used dataset by the graph matching community. It is composed by more than 140K graphs, generated according to different generation models (2D,3D,4D regular and nearly-regular meshes, bounded valence and randomly generated graphs).
The main issue of the MIVIA graphs dataset is nowadays its old age: the maximum size of the graphs is only 1000 nodes (evidently an appropriate size for the applications and the resources available at the date of the dataset creation). Furthermore, only very sparse graphs were included, being the maximum density fixed to 0.1, thus neglecting the possibility to evaluate the algorithms over more dense and of course more challenging graphs.
Nowadays, exact graph matching algorithms may be profitably used in those applications where the number of nodes is significantly higher: from thousands of nodes in biological applications (for representing protein structures) to tents of thousands of nodes in social networks (for representing the users and their relationships).
In order to face with the above considerations, a novel dataset, namely the MIVIA LDGraphs dataset, has been designed.
The MIVIA LDGraphs dataset
The computational cost of many graph matching algorithms (both in terms of time and space) is strongly related to the typology of graphs, and in particular to the following factors:
- the number of nodes of both target and pattern graphs
- the density of a graph (that is the ratio between the number of edges and the number of edges in a complete graph with the same number of nodes)
Thus, it is important to test the algorithms with the possibility to control such parameters so as to well understand in which situations an algorithm is better than another one. Considering that such control is not possible in real datasets, a synthetic dataset has been designed with a double aim: (1) the model used for the generation needs to be based on a small number of parameters, that can be easily combined so as to test under all the possible situations; (2) the model should not introduce any limitations on the graphs that could be exploited by special-case algorithms (for instance, polynomial algorithms exist for the case where the valence of the nodes is bounded by a constant independent of the graph size).
Starting from the above considerations, the graphs in the dataset have been generated with the Random Erdős–Rényi model (in particular, with the version proposed by Edgard Gilbert). The model requires the set up of two different parameters: the number of nodes N and the edge probability η (which determines the density of the graph), so that each of the possible edges is chosen with probability η and all the possible edges are equally probable.
Labeled and unlabeled graphs
Another very important factor which may impact on the cost of the matching is the label attached to each node, both in terms of number and distribution. Thus, it is also important to evaluate if and how a graph matching algorithm is able to exploit this kind of information.
The MIVIA LDGraphs dataset contains both labeled and unlabeled graphs. In particular, as for labeled graphs, two different labeled versions have been considered:
- uniform distribution: the labels are generated with a discrete uniform distribution in the interval [1,M], being M = 8.
- non uniform distribution: the labels are generated with a gaussian distribution centered in M/2 and truncated in the interval [1,M], being M = 8 as in the previous case.
Description
The MIVIA LDGraphs dataset is composed by a set of pairs of graphs, such that the first graph (the pattern graph) is a subgraph of the second graph (the target graph).
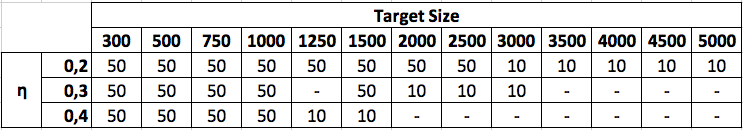
In more details, 2.550 unlabeled graphs have been first generated as target graphs by varying the setup parameters characteristic of the above mentioned typologies of graphs and by considering a set of dimensions (in terms of number of nodes) ranging from 300 to 10.000 nodes and a η value ranging from medium-low density values (0.2) to medium-high density values (0.4). For each setup, a number of graphs ranging between 10 and 50 graphs (depending on the graph size and typology) has been generated.
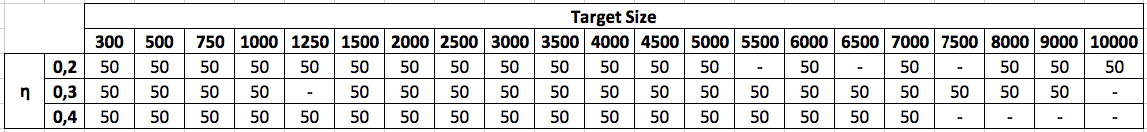
The maximum size of unlabeled graphs has been fixed to 5.000 nodes, while it is fixed to 10.000 for labeled graphs. Totally, 6.350 target graphs (including both labeled and unlabeled versions), and thus 6.350 pairs of graphs, have been finally obtained. For each target graph, a pattern subgraph whose size is 20% of the target graph has been derived. The nodes of the pattern graph have been randomly chosen, with the only constraint that the graph had to be connected.
In the following, two tables describing the datasets have been defined. In particular, η values (0.2, 0.3, 0.4) and target size (from 300 to 10.000) are reported on the rows and on the columns, respectively. The number in the generic cell (10 or 50) identifies the number of repetitions generated for that particular couple η – target size.
Unlabeled graphs
Labeled graphs (uniform and non uniform)
The dataset is available under request at the following link.
The MIVIA LDGraphs (MIVIA Large Dense Graphs) dataset is a new dataset for benchmarking exact graph matching algorithms. It aims to extend the MIVIA graphs dataset, widely used in the last ten years, with bigger and more dense graphs, so as to face with the problems nowadays encountered in real applications devoted for instance to bioinformatics and social network analysis.
If you are using this library please cite:
The MIVIA graphs dataset
The MIVIA graphs dataset has been one of the mostly used dataset by the graph matching community. It is composed by more than 140K graphs, generated according to different generation models (2D,3D,4D regular and nearly-regular meshes, bounded valence and randomly generated graphs).
The main issue of the MIVIA graphs dataset is nowadays its old age: the maximum size of the graphs is only 1000 nodes (evidently an appropriate size for the applications and the resources available at the date of the dataset creation). Furthermore, only very sparse graphs were included, being the maximum density fixed to 0.1, thus neglecting the possibility to evaluate the algorithms over more dense and of course more challenging graphs.
Nowadays, exact graph matching algorithms may be profitably used in those applications where the number of nodes is significantly higher: from thousands of nodes in biological applications (for representing protein structures) to tents of thousands of nodes in social networks (for representing the users and their relationships).
In order to face with the above considerations, a novel dataset, namely the MIVIA LDGraphs dataset, has been designed.
The MIVIA LDGraphs dataset
The computational cost of many graph matching algorithms (both in terms of time and space) is strongly related to the typology of graphs, and in particular to the following factors:
- the number of nodes of both target and pattern graphs
- the density of a graph (that is the ratio between the number of edges and the number of edges in a complete graph with the same number of nodes)
Thus, it is important to test the algorithms with the possibility to control such parameters so as to well understand in which situations an algorithm is better than another one. Considering that such control is not possible in real datasets, a synthetic dataset has been designed with a double aim: (1) the model used for the generation needs to be based on a small number of parameters, that can be easily combined so as to test under all the possible situations; (2) the model should not introduce any limitations on the graphs that could be exploited by special-case algorithms (for instance, polynomial algorithms exist for the case where the valence of the nodes is bounded by a constant independent of the graph size).
Starting from the above considerations, the graphs in the dataset have been generated with the Random Erdős–Rényi model (in particular, with the version proposed by Edgard Gilbert). The model requires the set up of two different parameters: the number of nodes N and the edge probability η (which determines the density of the graph), so that each of the possible edges is chosen with probability η and all the possible edges are equally probable.
Labeled and unlabeled graphs
Another very important factor which may impact on the cost of the matching is the label attached to each node, both in terms of number and distribution. Thus, it is also important to evaluate if and how a graph matching algorithm is able to exploit this kind of information.
The MIVIA LDGraphs dataset contains both labeled and unlabeled graphs. In particular, as for labeled graphs, two different labeled versions have been considered:
- uniform distribution: the labels are generated with a discrete uniform distribution in the interval [1,M], being M = 8.
- non uniform distribution: the labels are generated with a gaussian distribution centered in M/2 and truncated in the interval [1,M], being M = 8 as in the previous case.
Description
The MIVIA LDGraphs dataset is composed by a set of pairs of graphs, such that the first graph (the pattern graph) is a subgraph of the second graph (the target graph).
In more details, 2.550 unlabeled graphs have been first generated as target graphs by varying the setup parameters characteristic of the above mentioned typologies of graphs and by considering a set of dimensions (in terms of number of nodes) ranging from 300 to 10.000 nodes and a η value ranging from medium-low density values (0.2) to medium-high density values (0.4). For each setup, a number of graphs ranging between 10 and 50 graphs (depending on the graph size and typology) has been generated.
The maximum size of unlabeled graphs has been fixed to 5.000 nodes, while it is fixed to 10.000 for labeled graphs. Totally, 6.350 target graphs (including both labeled and unlabeled versions), and thus 6.350 pairs of graphs, have been finally obtained. For each target graph, a pattern subgraph whose size is 20% of the target graph has been derived. The nodes of the pattern graph have been randomly chosen, with the only constraint that the graph had to be connected.
In the following, two tables describing the datasets have been defined. In particular, η values (0.2, 0.3, 0.4) and target size (from 300 to 10.000) are reported on the rows and on the columns, respectively. The number in the generic cell (10 or 50) identifies the number of repetitions generated for that particular couple η – target size.
Unlabeled graphs
Labeled graphs (uniform and non uniform)

The dataset is available under request at the following link.